SARS-CoV-2 genomic sequences available from GISAID as of May 31st 2021
Published: 10 June 2021
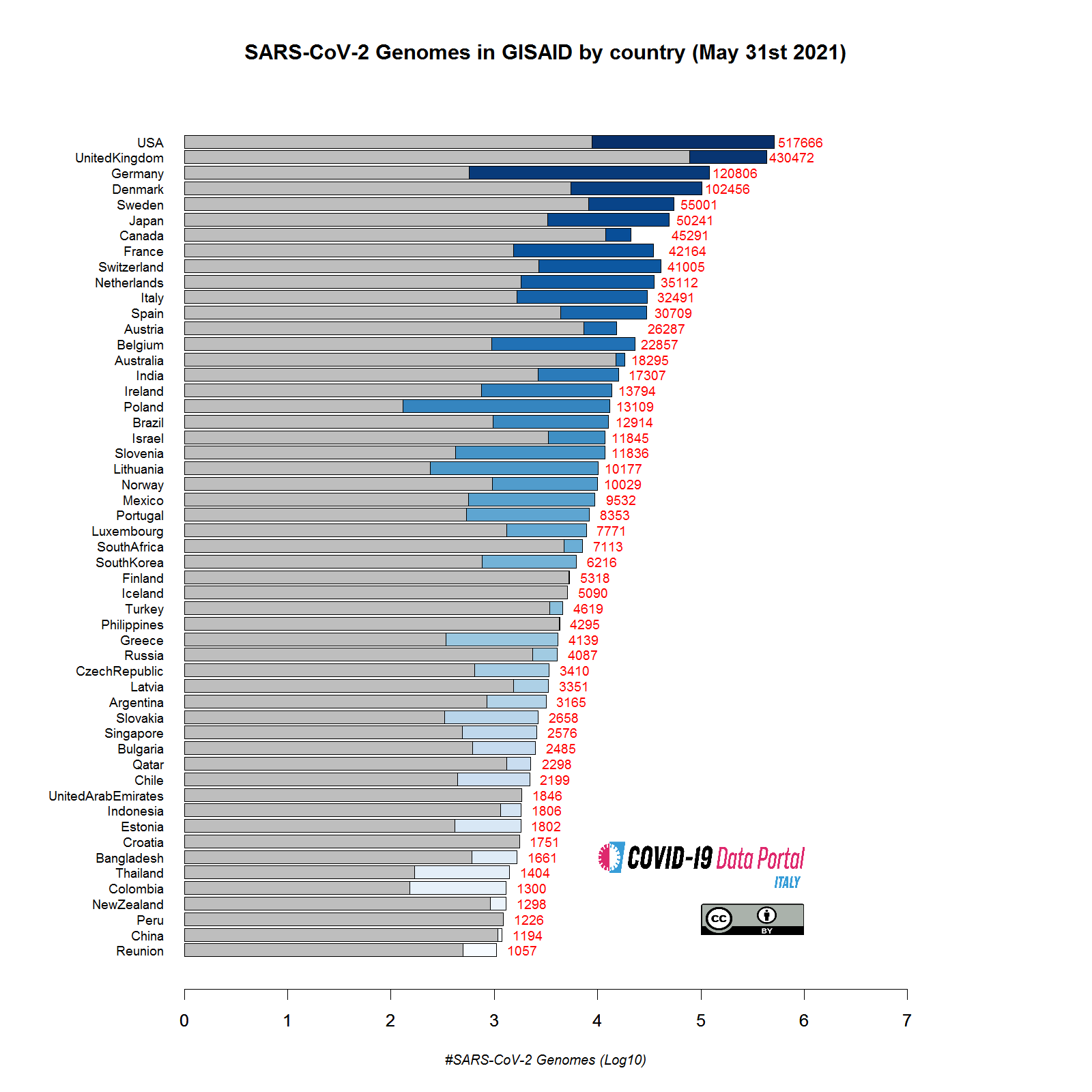
This figure provides an overview of the number of SARS-CoV-2 genomic sequences available in the GISAID database for different countries. Two colors are used to highlight:
- Gray: genomes already present in the database at the time of our previous update (Mar 31st).
- Blue: new genomes submitted in the last two months (from Apr 1st to May 31st).
Several countries, including Italy, have produced and submitted an increased number of “novel” sequences during the last two months. In some cases (see for example Poland), the number of genomic sequences is almost doubled with respect to the previous update. Overall the observed patterns indicate an increased capacity to produce and analyse pathogen genomic data. Indeed a further increase in the number of countries overcoming the threshold of 1000 sequenced genomes (minimal threshold to be included in this report), from 40 to 54 is also observed. Unsurprisingly, the number of sequences deposited by countries where the number of reported cases of COVID-19 is close to zero, such as China, Iceland and Israel, is very limited or even equal to zero.
The substantial increase in the number of genomes available for many countries reflects a clear improvement in “genomic surveillance” programs both at the national and international levels. The availability of an increasing amount of data will allow more careful monitoring of the evolution of the virus and facilitate a more rapid and precise identification of novel potentially dangerous variants.
Source: GISAID