Salient features of the new Omicron variant (B.1.1.529)
Published: 02 December 2021
Here we present a description of the main features of the Omicron variant, based on currently available data.
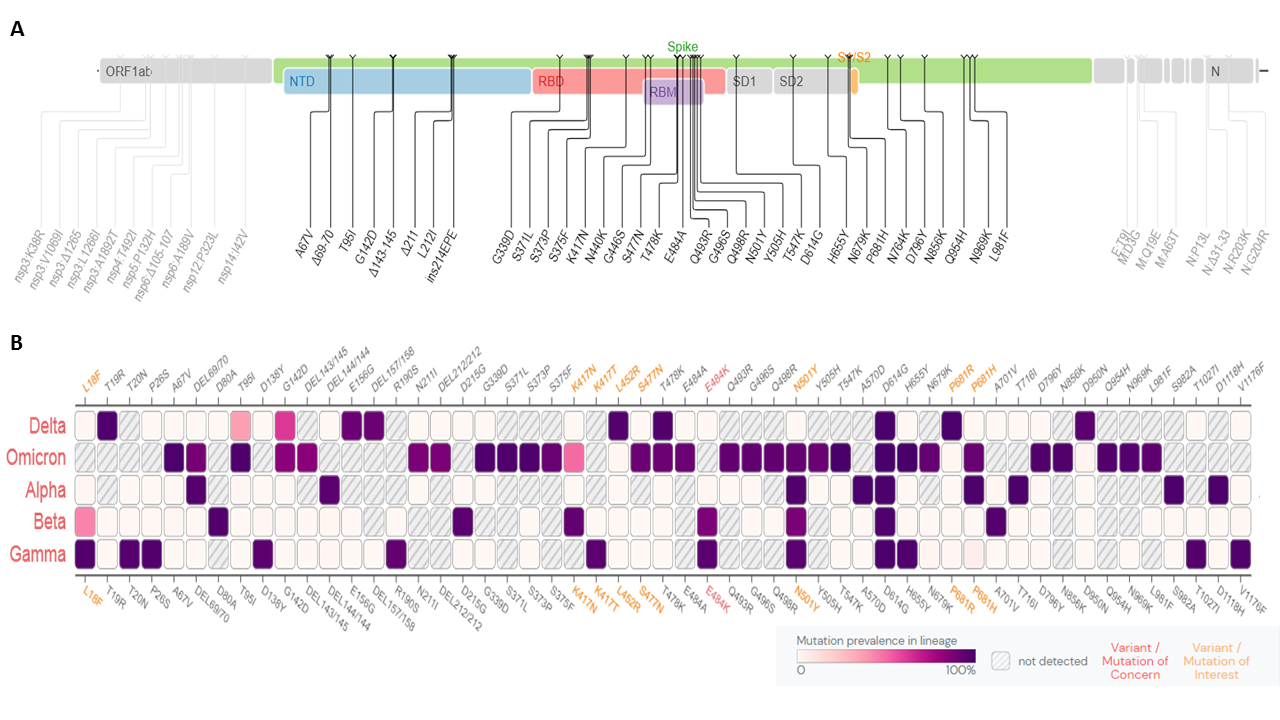
Panel A presents a schematic representation of the key mutations identified in the genome of B.1.1.529, specifically in the Spike glycoprotein; the scale is not proportional to the actual size of genomic features and emphasizes the size of the Spike gene.
Panel B provides a comparison between the characteristic mutations of the Omicron variant and the mutations identified in the other Variants of Concern (VOCs) of SARS-CoV-2 as defined by the World Health Organization. The aim of this comparison is to highlight similarities and differences between Omicron and other VOCs.
On November 26th 2021 the World Health Organization (WHO) designated the SARS-CoV-2 lineage known as B.1.1.529, isolated in South Africa at the beginning of the month, as a new Variant of Concern (VOC) named <<Omicron>>. This decision was based on the analyses run by the Technical Advisory Group on Virus Evolution (TAG-VE), which assessed that the mutations observed in the genome of the new variant could confer to the virus the ability to spread more rapidly, to cause a more severe form of COVID-19, or to be recognized in a less efficient manner by the immune system of vaccinated individuals. The Omicron variant carries an impressive number of mutations in its genome, at least 30 of these are localized in the Spike protein (panel A).
Some of these mutation have been already observed in other VOCs (an example is the deletion in position 69/70, which was previously identified in the Alpha variant, panel B); on the other hand Omicron also presents completely new mutations, which were never observed together in the same lineage and represent an additional source of concern. Preliminary studies highlight that some of these mutations may be associated with increased transmissibility [1], higher binding affinity for the ACE2 receptor [2], an enhanced immune escape [3], and finally an increase in viral load [4].
However, our current knowledge on Omicron is still very limited, and at the moment it is impossible to predict if the novel variant is going to have any impact on the epidemiology of COVID-19.
To close this gap, WHO is promoting a close collaboration between scientists worldwide, to study the transmissibility of Omicron, the severity of the infection caused by the novel variant (including symptoms) and the effectiveness of treatments, the performance of vaccines and diagnostic tests.
Preliminary data suggest that:
- It is not clear whether Omicron is more transmissible than other variants, especially Delta that is currently the most widespread variant worldwide. However, the number of people testing positive for COVID-19 is increasing in those regions of South Africa where the new variant is endemic, but further epidemiological studies are required to understand if this increase is caused by Omicron or by other factors.
- It is not clear if Omicron causes a more severe form of the disease, and currently available data do not suggest that symptoms caused by this variant are any different. The current increase of hospitalizations observed in South Africa may be the consequence of a general increase in the number of infected people, and also of low vaccination rate in that country (around 25% of the total population), rather than the result of specific infections with Omicron.
- Risk of being reinfected may be increased with Omicron with respect to other variants, however data are still limited. Furthermore, WHO is actively working to evaluate the potential impact of Omicron on the effectiveness of existing vaccines. Vaccines remain effective against severe disease and death.
- The widely used PCR tests used for diagnosis of COVID-19 are effective in detecting the Omicron variant too. Further studies are required to determine whether other testing methods, such as rapid antigen detection tests, are affected by Omicron.
- Corticosteroids and IL-6 Receptor Blockers are still effective for treating patients with severe COVID-19. Further analyses are required to assess if Omicron affects the efficacy of other treatments.
While this report briefly summarizes the state of the art, more information about the most important features of Omicron will become available in the following weeks. Constant monitoring of the spread of the novel variant through genomic surveillance, and additional studies for the characterization of the properties of Omicron will be required to understand its “real” epidemiological impact.
Additional information and updates about Omicron can be found at the following links: WHO, ECDC, Outbreak.info, CoVariants, Wikipedia.
Sources:
WHO (classification and state of the art),
CoVariants (genetic characteristics),
[1] “Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity”. S.Y. Gong et al. 11 September 2021,
[2] “SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution”. Zahradník, J., Marciano, S., Shemesh, M. et al. 16 August 2021,
[3] “Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy”. D. Benvenuto, S. Angeletti and M. Giovanetti et al. 10 April 2020,
[4] “Saudi Arabian SARS-CoV-2 genomes implicate a mutant Nucleocapsid protein in modulating host interactions and increased viral load in COVID-19 patients”. Mourier et al. 20 May 2021 (preprint),
Outbreak.info e Wikipedia (figures)