Prevalence of SARS-CoV-2 lineages of potential epidemiological concern
Published: 27 February 2021
A comparison between different time intervals is provided to illustrate the dynamics of circulation and or possible increases/decreases in the prevalence of these lineages. Table 1 and Table 2 report data as obtained from the analysis of genomic sequences obtained between Jan 27th and Feb 26th, and between Dec 27th and Jan 26th respectively. Table 3 compares the prevalence of these lineages in different Italian regions in the last month (Jan 27th and Feb 26th), with the “global” prevalence, as measured on all currently available genomes. This was required as unfortunately, the number of available genomic sequences is still limited form some regions in our country.
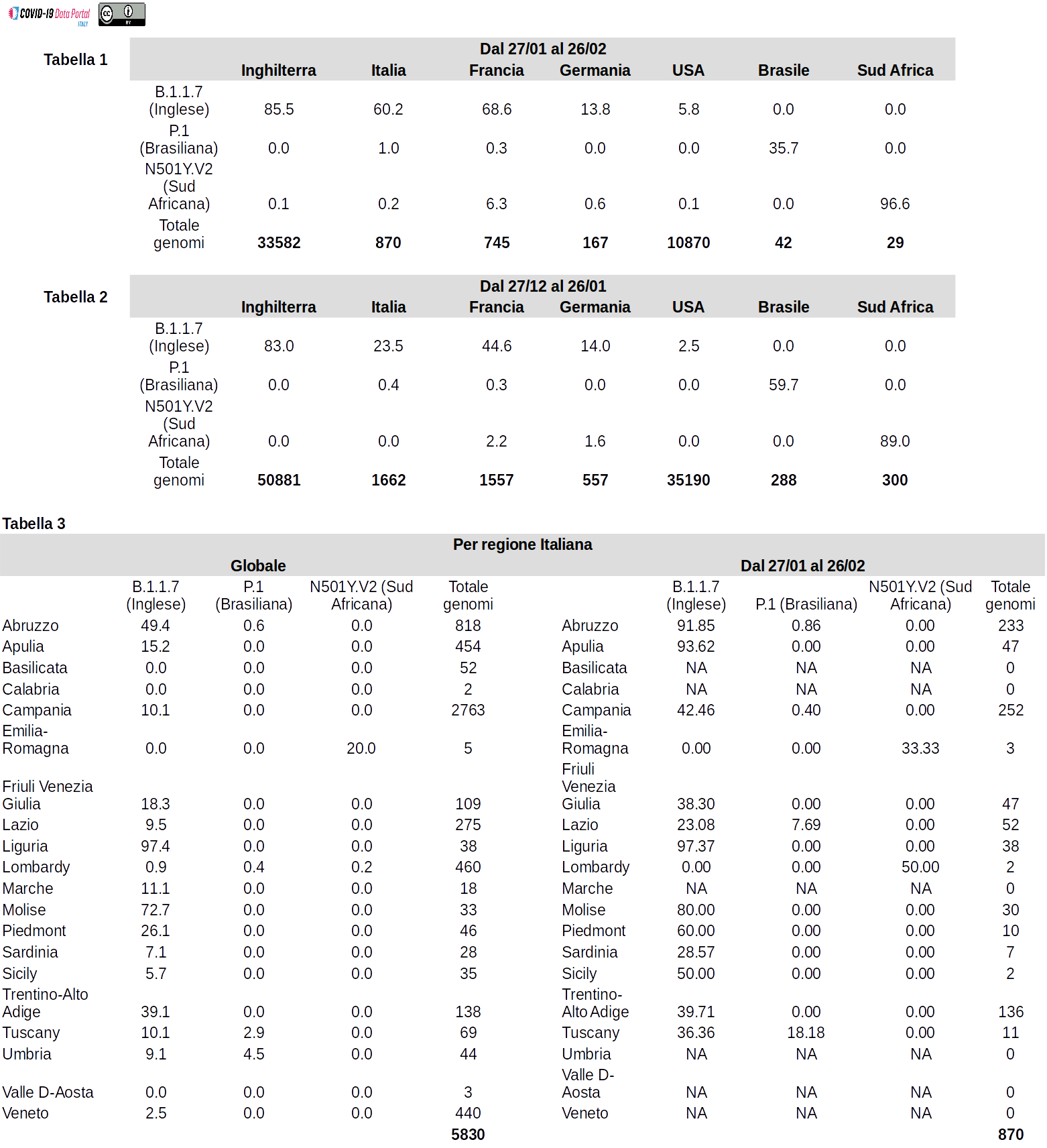
The tables report the prevalence (proportion of all the available genomes) known as the “English (B.1.1.7)”, “South African (501Y.V2)” and “Brazilian (P.1)” variants, that are considered to represent a potential risk and are under scrutiny by national and supranational health authorities Table 1 reports the prevalence of the 3 lineages in the last month (between 27/01/2021 and 26/02/2021) in different countries. Table 2 shows equivalent data but for the previous month (between 27/12/200 and 26/01/2021). In Table 1 and Table 2 the last row is used to indicate the total numbers of genomic sequences considered in our analyses. Table 3 reports the prevalence of the above-mentioned lineages in different Italian regions. Prevalence was calculated both by considering all the available genomic sequences (Global) and/or only genomes deposited in a public database between 27/01/2021 and 26/02/2021 (From 27/01 to 26/02). The column “Total genomes” reports the total number of genomes considered.
IMPORTANT: Since available data are fragmented and not homogeneous between different Italian regions, the information reported in this analysis is purely descriptive, and does not necessarily represent the real situation in our country.
IMPORTANT: while these are up to date, our analyses cannot offer a complete overview of the current situation in our country. Available data are fragmented and do not have the same level of completeness or accuracy for different geographic regions. The information reported in this analysis is purely descriptive and does not necessarily represent the real situation in Italy.
In other words, it is like looking at a very low-res picture: details are blurred and it is nearly impossible to understand the real situation with a sufficient degree of precision. Improving the resolution of this picture both at the spatial and temporal level, that is increasing the number of viral genomes regularly sequenced and shared in public repositories in our country, is one of the most ambitious objectives of this portal.
Variants of potential epidemiological relevance
English Variant (lineage B.1.1.7)
B.1.1.7 was first detected in the South of the UK in mid-December 2020. This lineage is currently the predominant type of SARS-CoV-2 in the UK. The incidence of COVID-19 cases in England increased from early December 2020 until reaching a peak in early January 2021; A decreasing trend was observed from January 11th to 24th, likely due to the implementation of rigorous health control measures and social distancing.
Similar dynamics were also recorded in Denmark, Ireland and the Netherlands, where this lineage was subsequently isolated. In the UK and Northern Ireland, B.1.1.7 has been shown to have higher transmissibility. Moreover preliminary observations suggest also a greater severity of the disease, although further experimental tests and clinical investigations are required [4].
At the time of writing, 958 genomic sequences associated with the B.1.1.7 lineage (out of a total of 5830) have been identified and deposited in the GISAID database from Italy. All the specimens associated with these genome sequences have been isolated after 14/12/2020. By direct comparison between Table 1 and Table 2, it is possible to observe that the prevalence of this viral lineage seems to have increased significantly in our country between January and February (from 23.5 to 60.2% of cases). The same trend is observed in France. While in the UK, B.1.1.7 has clearly become the most prevalent variant, as early as January. B.1.1.7 shows only a reduced prevalence in non-European countries such as the United States of America, Brazil or South Africa.
From Table 3 it is possible to observe that, according to currently available data, the distribution of B.1.1.7 does not seem to be uniform between different Italian regions. However, it should be stressed that currently available data were not collected in a uniform and homogeneous manner (see 0 and/or missing values "NA"). For this reason, it is not currently possible to obtain an accurate estimate of the prevalence of this, or any other lineage in different Italian regions, based solely on genome sequencing data.
Additionally, it can not be excluded that the increase in the number of viral genomes of this type might be due, at least in part, to the growing attention that has been registered in the last 2 months for this variant at the epidemiological level, but also by the media. This may have led to an overestimate of its prevalence. A consideration that applies in particular to those Italian regions in which B.1.1.7 seems to reach a prevalence higher than 90%.
South African variant (lineage 501Y.V2)
The 501Y.V2 lineage was first identified in South Africa in December 2020, where it is currently the most widespread viral type. As of January 25, 2021, it has been reported in 31 countries. In South Africa, weekly cases have increased since early November and reached a peak in early January. In the last two weeks, the trend has been decreasing. Preliminary data indicate that this lineage may also be characterized by greater transmissibility; while it is currently unclear whether it is associated with more severe clinical manifestations.
For Italy, only 2 genomic sequences of this viral type are currently deposited in the GISAID database. In light of this very reduced number, no consideration can be made on the prevalence of 501Y.V2 in our country at present.
As can be seen from Table 1 and Table 2 this variant is associated with a very significant proportion (~ 90%) of the SARS-CoV-2 genomes sequenced in South Africa but does not reach a high prevalence in the other countries herein considered.
Brazilian variant (lineage P.1)
The P.1 lineage was first reported in Japan on 10/01/2021, where it was isolated in 4 travellers arriving from Brazil, and subsequently identified also in South Korea, again in travellers from Brazil. As of January 25th 2021, the P.1 lineage was reported in 8 countries, including Italy. In Brazil, the number of new weekly cases in the last two weeks is reported to be higher compared to September-November 2020, and COVID-19 related deaths have increased since the beginning of November. Preliminary investigations in Manaus, Amazonas state, report an increase in the proportion of COVOD-19 cases associated with P.1, from 52.2% (35/67) in December 2020 to 85.4% (41/48) in January 2021. Highlighting ongoing local transmission and suggesting potential greater transmissibility or propensity for reinfection, although available data are limited. There is no evidence of increased severity of the disease.
These data are in line with those reported in Table 2. While according to currently available data, as reported in Table 1, the prevalence of P.1 seems to be decreasing in January-February 2021. However, it should be noted that these estimates are based on a small sample (only 38 genomes).
At present, there are only 12 sequences of this viral type in the GISAID database for our country. Also, in this case, any consideration on the prevalence of this variant in Italy would be premature.
Source: https://www.gisaid.org/ (genomic data)
Source: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (epidemiological data)
Please see https://cov-lineages.org/global_report.html for daily updates on the prevalence of SARS-CoV-2 lineages worldwide
Bibliography
[1] “Weekly epidemiological update -27January 2021” (https://www.who.int/publications/m/item/weekly-epidemiological-update—27-january-2021)
[2] “European Centre for Disease Prevention and Control. Risk related to spread of new SARS-CoV-2 variants of concern in the EU/EEA, first update –21 January 2021. ECDC: Stockholm; 2021.” (https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-risk-related-to-spread-of-new-SARS-CoV-2-variants-EU-EEA-first-update.pdf)
[3] “European Centre for Disease Prevention and Control. Sequencing of SARS-CoV-2: first update. 18 January 2021. ECDC: Stockholm; 2021. (https://www.ecdc.europa.eu/sites/default/files/documents/Sequencing-of-SARS-CoV-2-first-update.pdf)
[4] NERVTAG paper on COVID-19 variant of concern B.1.1.7. The Government of the United Kingdom of Great Britain and Northern Ireland; 2021. Available at: https://www.gov.uk/government/publications/nervtag-paper-on-covid-19-variant-of-concern-b117